publications
2025
-
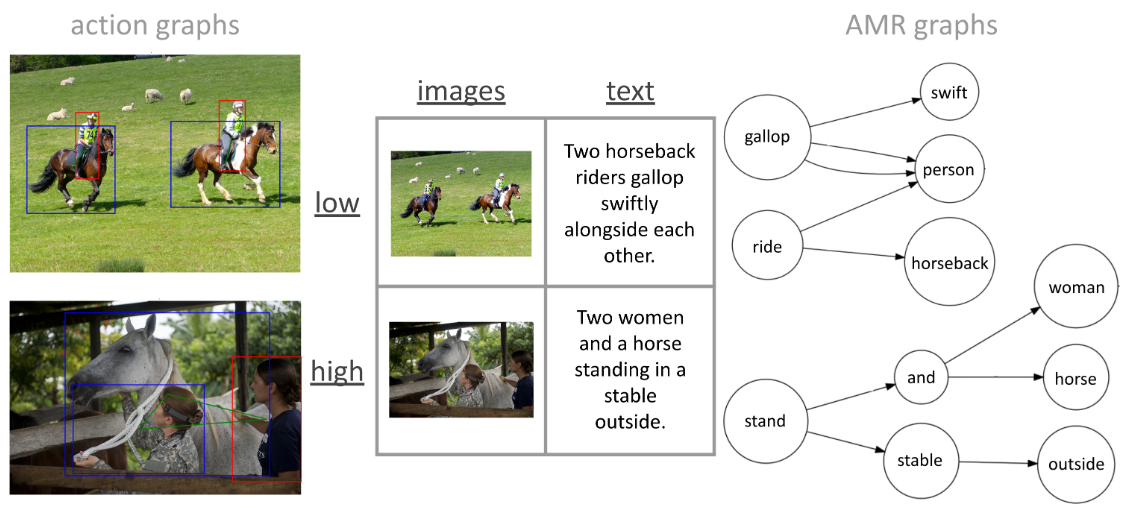 Helena Balabin, Antonietta Gabriella Liuzzi, Kevin Statz, and 3 more authorsNeurobiology of Language, Jul 2025[Stage 1 In-Principle Acceptance: Registered Report]
Helena Balabin, Antonietta Gabriella Liuzzi, Kevin Statz, and 3 more authorsNeurobiology of Language, Jul 2025[Stage 1 In-Principle Acceptance: Registered Report] -
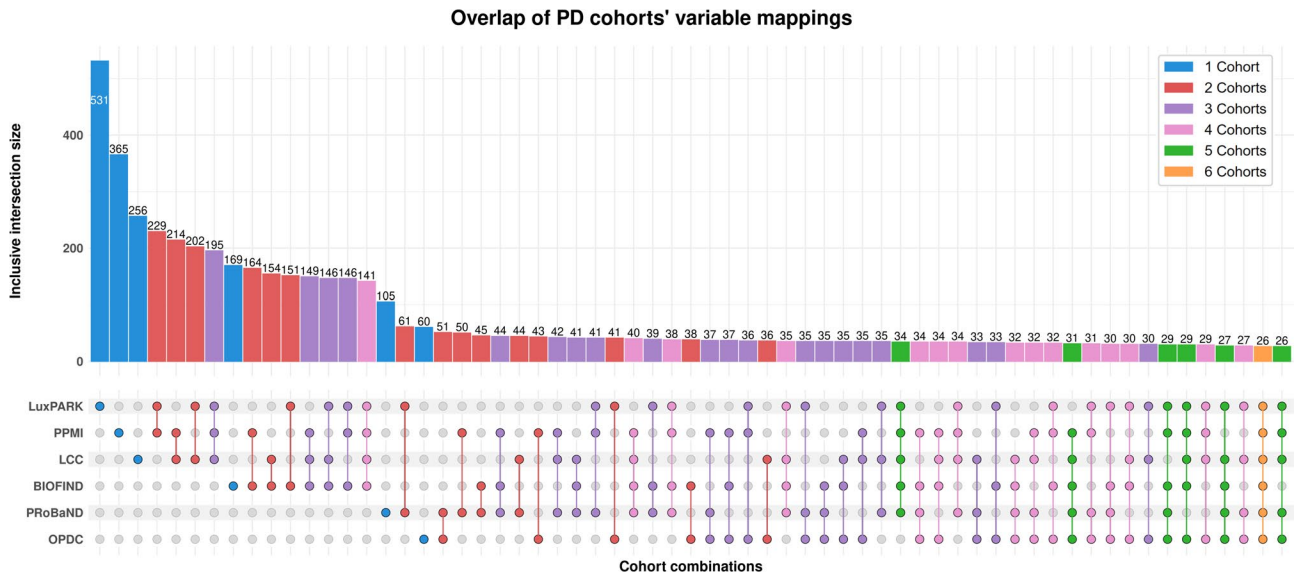 Yasamin Salimi , Tim Adams, Mehmet Can Ay, and 3 more authorsScientific Reports, Jun 2025
Yasamin Salimi , Tim Adams, Mehmet Can Ay, and 3 more authorsScientific Reports, Jun 2025Data Harmonization is an important yet time-consuming process. With the recent popularity of applications using Language Models (LMs) due to their high capabilities in text understanding, we investigated whether LMs could facilitate data harmonization for clinical use cases. To evaluate this, we created PASSIONATE, a novel Parkinson’s disease (PD) variable mapping schema as a ground truth source for pairwise cohort harmonization using LLMs. Additionally, we extended our investigation using an existing Alzheimer’s disease (AD) CDM. We computed text embeddings based on two language models to perform automated cohort harmonization for both AD and PD. We additionally compared the results to a baseline method using fuzzy string matching to determine the degree to which the semantic capabilities of language models can be utilized for automated cohort harmonization. We found that mappings based on text embeddings performed significantly better than those generated by fuzzy string matching, reaching an average accuracy of over 80% for almost all tested PD cohorts. When extended to a further neighborhood of possible matches, the accuracy could be improved to up to 96%. Our results suggest that language models can be used for automated harmonization with a high accuracy that can potentially be improved in the future by applying domain-trained models.
-
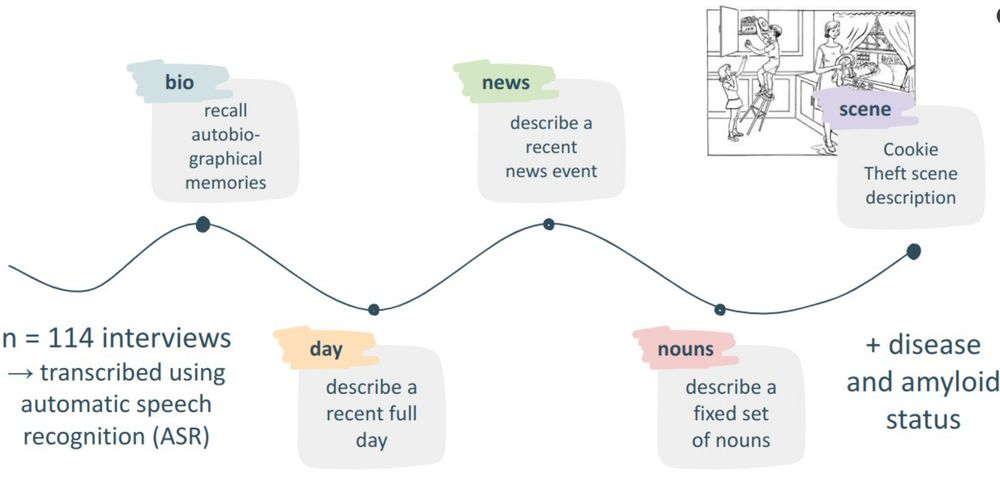 Helena Balabin, Bastiaan Tamm, Laure Spruyt, and 9 more authorsAlzheimer’s & Dementia, Jan 2025
Helena Balabin, Bastiaan Tamm, Laure Spruyt, and 9 more authorsAlzheimer’s & Dementia, Jan 2025INTRODUCTION The automated analysis of connected speech using natural language processing (NLP) emerges as a possible biomarker for Alzheimer’s disease (AD). However, it remains unclear which types of connected speech are most sensitive and specific for the detection of AD. METHODS We applied a language model to automatically transcribed connected speech from 114 Flemish-speaking individuals to first distinguish early AD patients from amyloid negative cognitively unimpaired (CU) and then amyloid negative from amyloid positive CU individuals using five different types of connected speech. RESULTS The language model was able to distinguish between amyloid negative CU subjects and AD patients with up to 81.9% sensitivity and 81.8% specificity. Discrimination between amyloid positive and negative CU individuals was less accurate, with up to 82.7% sensitivity and 74.0% specificity. Moreover, autobiographical interviews consistently outperformed scene descriptions. DISCUSSION Our findings highlight the value of autobiographical interviews for the automated analysis of connecting speech. Highlights This study compared five types of connected speech for the detection of early Alzheimer’s disease (AD). Autobiographical interviews yielded a higher specificity than scene descriptions. A preceding clinical AD classification task can refine the performance of amyloid status classification in cognitively healthy individuals.
2024
-
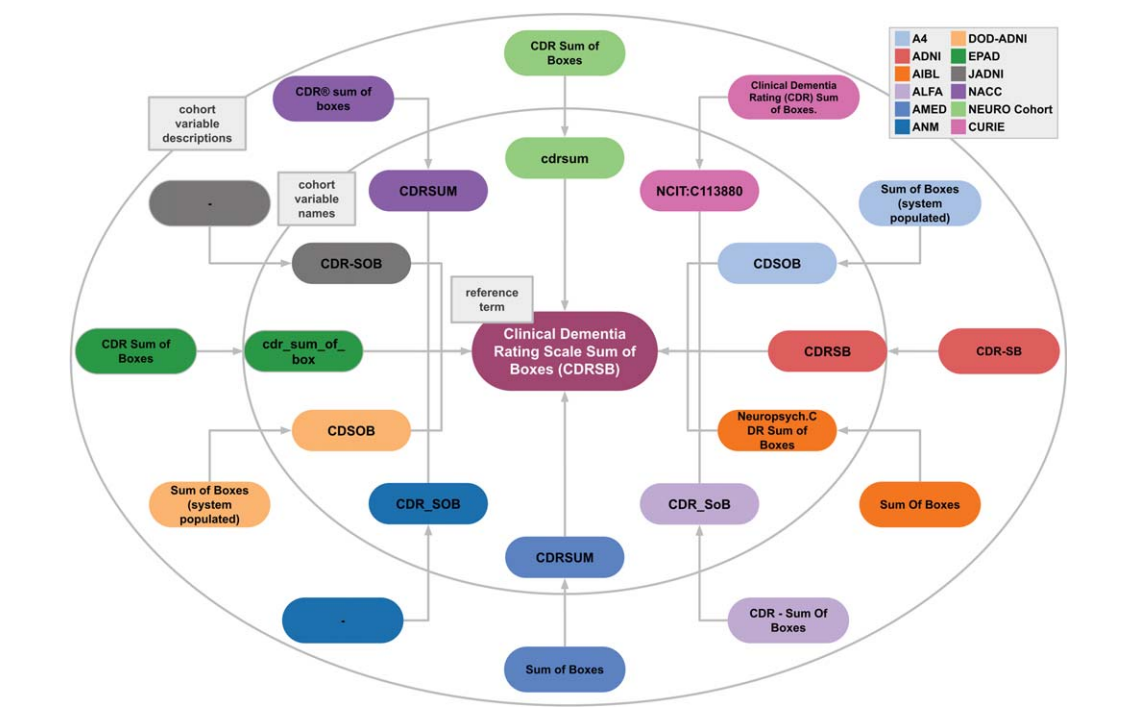 Philipp Wegner, Helena Balabin, Mehmet Can Ay, and 5 more authorsJournal of Alzheimer’s Disease, Apr 2024
Philipp Wegner, Helena Balabin, Mehmet Can Ay, and 5 more authorsJournal of Alzheimer’s Disease, Apr 2024 -
 Helena Balabin, Laure Spruyt, Ella Eycken, and 6 more authorsIn Proceedings of the First Workshop on Artificial Intelligence for Brain Encoding and Decoding at the AAAI Conference on Artificial Intelligence , Feb 2024
Helena Balabin, Laure Spruyt, Ella Eycken, and 6 more authorsIn Proceedings of the First Workshop on Artificial Intelligence for Brain Encoding and Decoding at the AAAI Conference on Artificial Intelligence , Feb 2024Early diagnosis of neurological disorders is a fast developing field of applied artificial intelligence (AI). In this context, AI- based language models have been increasingly used to distinguish cognitively healthy individuals from those affected by Alzheimer’s disease (AD) based on their connected speech. Yet, it remains unknown how the adaptation of the language models to the language and domain of the connected speech samples impacts the classification results. Here, we construct several classification tasks from Dutch Flemish samples of connected speech from a cohort of 74 healthy controls and 20 subjects affected by AD. First, we compare the classification performance of Dutch and multilingual models as well as models that incorporate long-range context. Additionally, we examine how varying amounts of fine-tuning data from a separate corpus of speech samples affect domain adaptation. We demonstrate that increasing fine-tuning data leads to increased domain adaptation, but it does not necessarily translate into improved classification performance. Furthermore, our findings support the use of language-specific models over multilingual ones, even for multilingual models that were trained to incorporate wider context.
2023
-
 Helena Balabin, Antonietta Gabriella Liuzzi, Jingyuan Sun, and 3 more authorsIn Proceedings of the 26th European Conference on Artificial Intelligence , Oct 2023
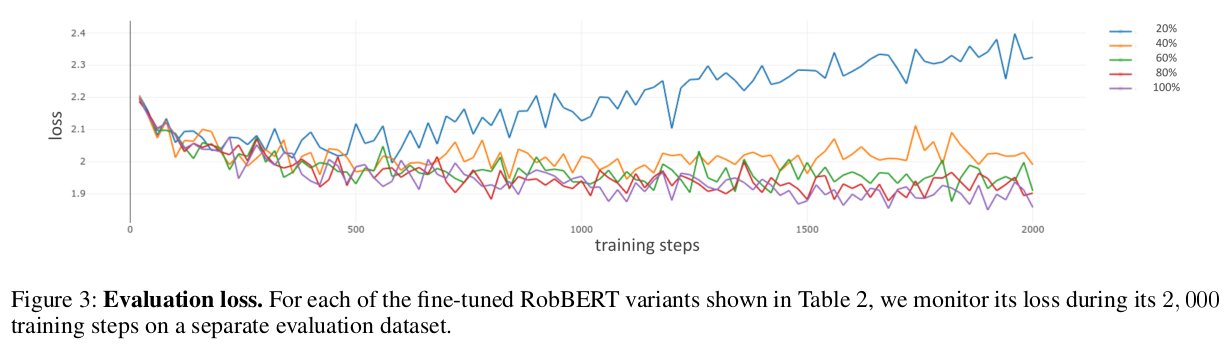
Helena Balabin, Antonietta Gabriella Liuzzi, Jingyuan Sun, and 3 more authorsIn Proceedings of the 26th European Conference on Artificial Intelligence , Oct 2023In recent years, representations from brain activity patterns and pre-trained language models have been linked to each other based on neural fits to validate hypotheses about language processing. Nonetheless, open questions remain about what intrinsic properties of language processing these neural fits reflect and whether they differ across neural fit approaches, brain networks, and models. In this study, we use parallel sentence and functional magnetic resonance imaging data to perform a comprehensive analysis of four paradigms (masked language modeling, pragmatic coherence, semantic comparison, and contrastive learning) representing linguistic hypotheses about sentence processing. We include three sentence embedding models for each paradigm, resulting in a total of 12 models, and examine differences in their neural fit to four different brain networks using regression-based neural encoding and Representational Similarity Analysis (RSA). Among the different models tested, GPT-2, SkipThoughts, and S-RoBERTa yielded the strongest correlations with language network patterns, whereas contrastive learning-based models resulted in overall low neural fits. Our findings demonstrate that neural fits vary across brain networks and models representing the same linguistic hypothesis (e.g., GPT-2 and GPT-3). More importantly, we show the need for both neural encoding and RSA as complementary methods to provide full understanding of neural fits. All code used in the analysis is publicly available: https://github.com/lcn-kul/sentencefmricomparison.
2022
-
 Vladimir Araujo, Helena Balabin, Julio Hurtado, and 2 more authorsIn Proceedings of the 2nd Conference of the Asia-Pacific Chapter of the Association for Computational Linguistics and the 12th International Joint Conference on Natural Language Processing , Nov 2022
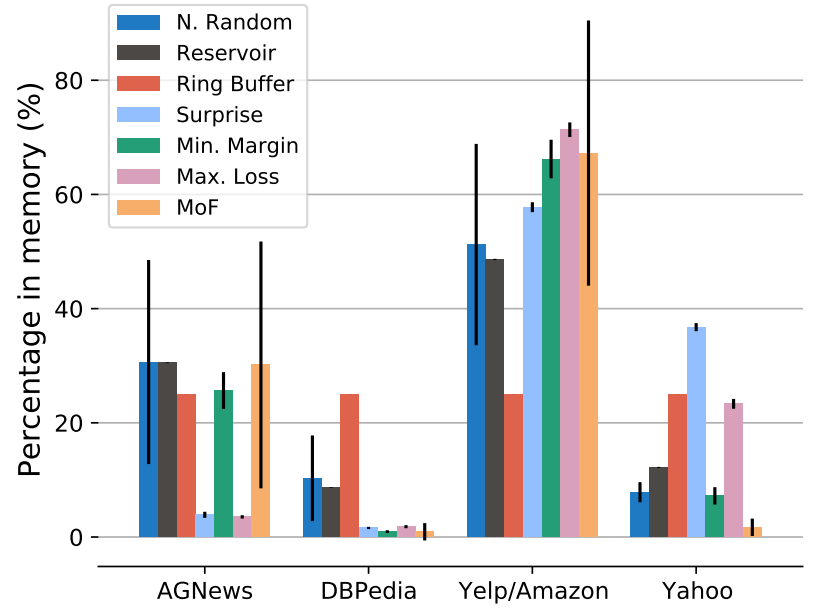
Vladimir Araujo, Helena Balabin, Julio Hurtado, and 2 more authorsIn Proceedings of the 2nd Conference of the Asia-Pacific Chapter of the Association for Computational Linguistics and the 12th International Joint Conference on Natural Language Processing , Nov 2022Lifelong language learning seeks to have models continuously learn multiple tasks in a sequential order without suffering from catastrophic forgetting. State-of-the-art approaches rely on sparse experience replay as the primary approach to prevent forgetting. Experience replay usually adopts sampling methods for the memory population; however, the effect of the chosen sampling strategy on model performance has not yet been studied. In this paper, we investigate how relevant the selective memory population is in the lifelong learning process of text classification and question-answering tasks. We found that methods that randomly store a uniform number of samples from the entire data stream lead to high performances, especially for low memory size, which is consistent with computer vision studies.
-
 Bastiaan Tamm, Helena Balabin, Rik Vandenberghe, and 1 more authorIn Interspeech 2022 , Oct 2022
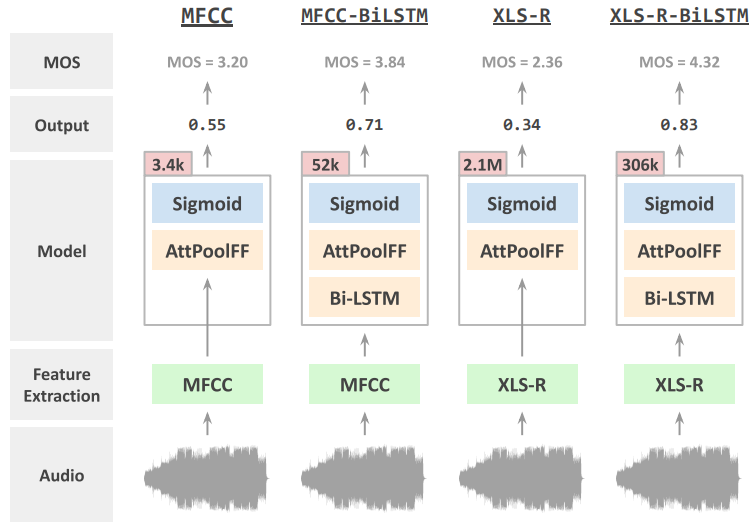
Bastiaan Tamm, Helena Balabin, Rik Vandenberghe, and 1 more authorIn Interspeech 2022 , Oct 2022Speech quality in online conferencing applications is typically assessed through human judgements in the form of the mean opinion score (MOS) metric. Since such a labor-intensive approach is not feasible for large-scale speech quality assessments in most settings, the focus has shifted towards automated MOS prediction through end-to-end training of deep neural networks (DNN). Instead of training a network from scratch, we propose to leverage the speech representations from the pre-trained wav2vec-based XLS-R model. However, the number of parameters of such a model exceeds task-specific DNNs by several orders of magnitude, which poses a challenge for resulting fine-tuning procedures on smaller datasets. Therefore, we opt to use pre-trained speech representations from XLS-R in a feature extraction rather than a fine-tuning setting, thereby significantly reducing the number of trainable model parameters. We compare our proposed XLS-R-based feature extractor to a Mel-frequency cepstral coefficient (MFCC)-based one, and experiment with various combinations of bidirectional long short term memory (Bi-LSTM) and attention pooling feedforward (AttPoolFF) networks trained on the output of the feature extractors. We demonstrate the increased performance of pre-trained XLS-R embeddings in terms a reduced root mean squared error (RMSE) on the ConferencingSpeech 2022 MOS prediction task.
-
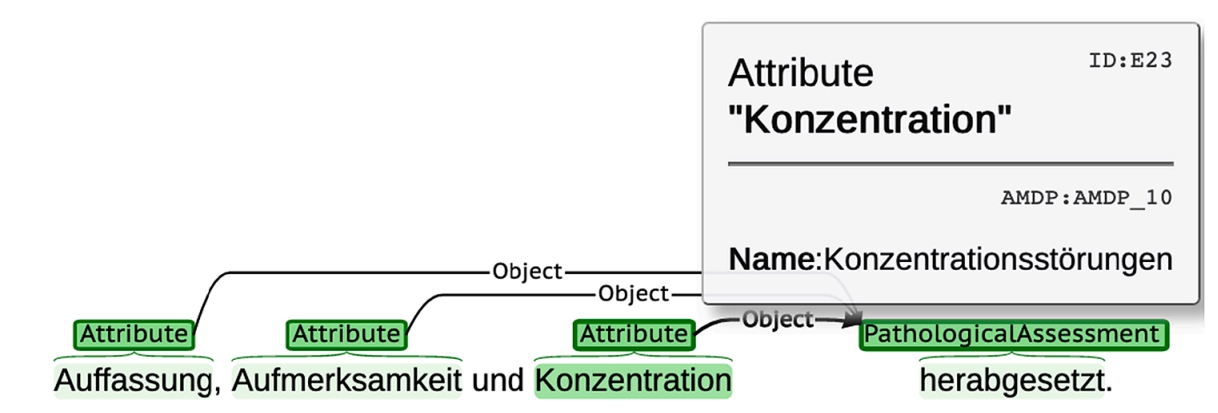 Sumit Madan, Fabian Julius Zimmer, Helena Balabin, and 7 more authorsInternational Journal of Medical Informatics, May 2022
Sumit Madan, Fabian Julius Zimmer, Helena Balabin, and 7 more authorsInternational Journal of Medical Informatics, May 2022Background Health care records provide large amounts of data with real-world and longitudinal aspects, which is advantageous for predictive analyses and improvements in personalized medicine. Text-based records are a main source of information in mental health. Therefore, application of text mining to the electronic health records – especially mental state examination – is a key approach for detection of psychiatric disease phenotypes that relate to treatment outcomes. Methods We focused on the mental state examination (MSE) in the patients’ discharge summaries as the key part of the psychiatric records. We prepared a sample of 150 text documents that we manually annotated for psychiatric attributes and symptoms. These documents were further divided into training and test sets. We designed and implemented a system to detect the psychiatric attributes automatically and linked the pathologically assessed attributes to AMDP terminology. This workflow uses a pre-trained neural network model, which is fine-tuned on the training set, and validated on the independent test set. Furthermore, a traditional NLP and rule-based component linked the recognized mentions to AMDP terminology. In a further step, we applied the system on a larger clinical dataset of 510 patients to extract their symptoms. Results The system identified the psychiatric attributes as well as their assessment (normal and pathological) and linked these entities to the AMDP terminology with an F1-score of 86% and 91% on an independent test set, respectively. Conclusion The development of the current text mining system and the results highlight the feasibility of text mining methods applied to MSE in electronic mental health care reports. Our findings pave the way for the secondary use of routine data in the field of mental health, facilitating further clinical data analyses.
-
 Helena Balabin, Charles Tapley Hoyt, Colin Birkenbihl, and 6 more authorsBioinformatics, Mar 2022
Helena Balabin, Charles Tapley Hoyt, Colin Birkenbihl, and 6 more authorsBioinformatics, Mar 2022The majority of biomedical knowledge is stored in structured databases or as unstructured text in scientific publications. This vast amount of information has led to numerous machine learning-based biological applications using either text through natural language processing (NLP) or structured data through knowledge graph embedding models. However, representations based on a single modality are inherently limited.To generate better representations of biological knowledge, we propose STonKGs, a Sophisticated Transformer trained on biomedical text and Knowledge Graphs (KGs). This multimodal Transformer uses combined input sequences of structured information from KGs and unstructured text data from biomedical literature to learn joint representations in a shared embedding space. First, we pre-trained STonKGs on a knowledge base assembled by the Integrated Network and Dynamical Reasoning Assembler consisting of millions of text-triple pairs extracted from biomedical literature by multiple NLP systems. Then, we benchmarked STonKGs against three baseline models trained on either one of the modalities (i.e. text or KG) across eight different classification tasks, each corresponding to a different biological application. Our results demonstrate that STonKGs outperforms both baselines, especially on the more challenging tasks with respect to the number of classes, improving upon the F1-score of the best baseline by up to 0.084 (i.e. from 0.881 to 0.965). Finally, our pre-trained model as well as the model architecture can be adapted to various other transfer learning applications.We make the source code and the Python package of STonKGs available at GitHub (https://github.com/stonkgs/stonkgs) and PyPI (https://pypi.org/project/stonkgs/). The pre-trained STonKGs models and the task-specific classification models are respectively available at https://huggingface.co/stonkgs/stonkgs-150k and https://zenodo.org/communities/stonkgs.Supplementary data are available at Bioinformatics online.
-
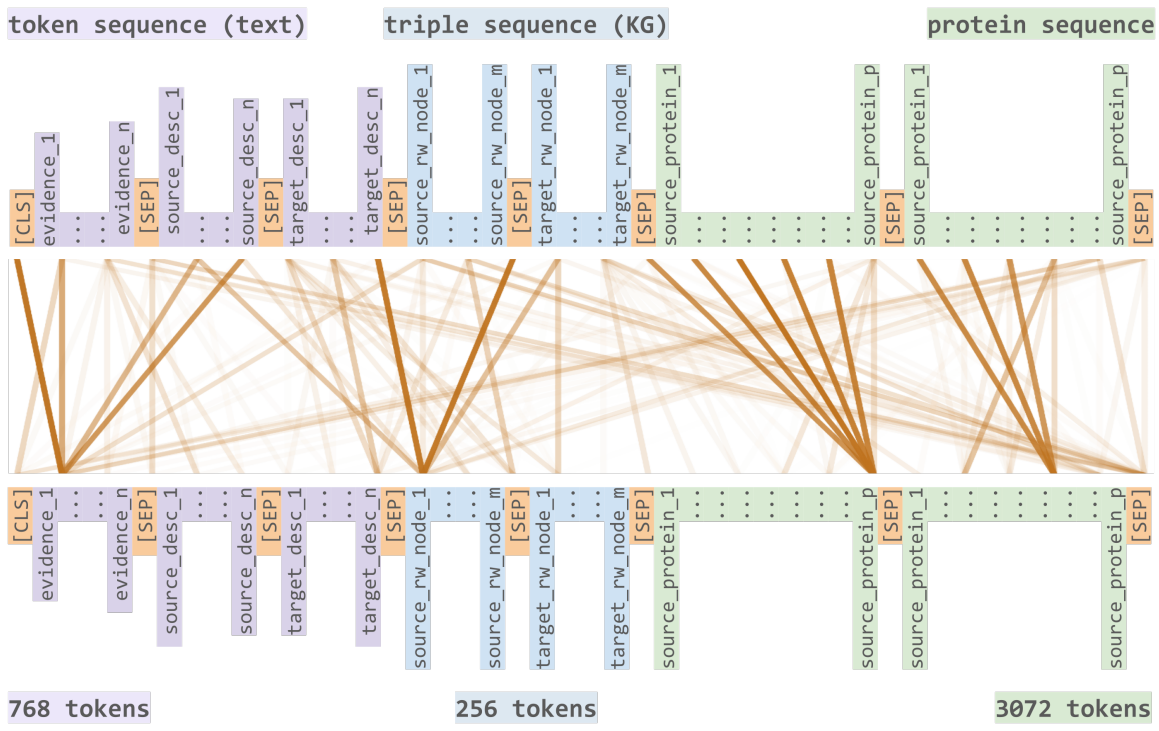 Helena Balabin, Charles Tapley Hoyt, Benjamin M Gyori, and 4 more authorsIn Proceedings of the 13th International Conference on Semantic Web Applications and Tools for Life Sciences , Jan 2022
Helena Balabin, Charles Tapley Hoyt, Benjamin M Gyori, and 4 more authorsIn Proceedings of the 13th International Conference on Semantic Web Applications and Tools for Life Sciences , Jan 2022While most approaches individually exploit unstructured data from the biomedical literature or structured data from biomedical knowledge graphs, their union can better exploit the advantages of such approaches, ultimately improving representations of biology. Using multimodal transformers for such purposes can improve performance on context dependent classification tasks, as demonstrated by our previous model, the Sophisticated Transformer Trained on Biomedical Text and Knowledge Graphs (STonKGs). In this work, we introduce Prot-STonKGs, a transformer aimed at learning all-encompassing representa- tions of protein-protein interactions. ProtSTonKGs presents an extension to our previous work by adding textual protein descriptions and amino acid sequences (i.e., structural information) to the text- and knowledge graph-based input sequence used in STonKGs. We benchmark Prot-STonKGs against STonKGs, resulting in improved F 1 scores by up to 0.066 (i.e., from 0.204 to 0.270) in several tasks such as predicting protein interactions in several contexts. Our work demonstrates how multimodal transformers can be used to integrate heterogeneous sources of information, paving the foundation for future approaches that use multiple modalities for biomedical applications.
2020
-
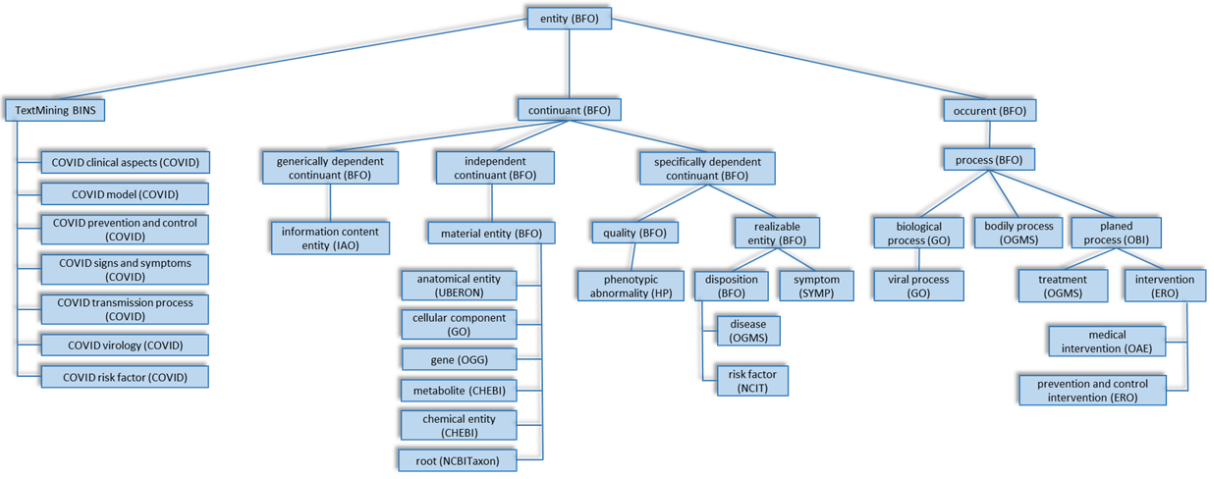 Astghik Sargsyan, Alpha Tom Kodamullil, Shounak Baksi, and 10 more authorsBioinformatics, Dec 2020
Astghik Sargsyan, Alpha Tom Kodamullil, Shounak Baksi, and 10 more authorsBioinformatics, Dec 2020The COVID-19 pandemic has prompted an impressive, worldwide response by the academic community. In order to support text mining approaches as well as data description, linking and harmonization in the context of COVID-19, we have developed an ontology representing major novel coronavirus (SARS-CoV-2) entities. The ontology has a strong scope on chemical entities suited for drug repurposing, as this is a major target of ongoing COVID-19 therapeutic development.The ontology comprises 2270 classes of concepts and 38 987 axioms (2622 logical axioms and 2434 declaration axioms). It depicts the roles of molecular and cellular entities in virus-host interactions and in the virus life cycle, as well as a wide spectrum of medical and epidemiological concepts linked to COVID-19. The performance of the ontology has been tested on Medline and the COVID-19 corpus provided by the Allen Institute.COVID-19 Ontology is released under a Creative Commons 4.0 License and shared via https://github.com/covid-19-ontology/covid-19. The ontology is also deposited in BioPortal at https://bioportal.bioontology.org/ontologies/COVID-19.Supplementary data are available at Bioinformatics online.